|
Leffingwell &
Associates
|
Services
and Software for the Perfume, Flavor, Food and
Beverage Industries
|
|
|

Alchemist WebPick Awarded by
the webzine of ChemWeb.com
|
Press Release - Leffingwell & Associates
- February 14, 2002, Canton, GA announced the public
release of theoretical 3-D Models of selected Human Olfactory
receptors and a rapid and simple methodology for determining the
putative odorant binding cavity.
Leffingwell & Associates provides services and software for
the perfume, flavor, food and beverage Industries.
Olfactory Receptor Protein 3-D Models and
Predicted Odorant Binding site Cavities
Leffingwell & Associates in a study of the predicted ligand
binding sites for certain Human OR's of chromosone 1, have found that
the largest cavity (as derived by CastP1-4)
in the complimentary regions falls in the transmembrane domains TM
3–7, especially in the region of TM3-TM5-TM6. This is
depicted below for selected Human Olfactory receptors as modeled by
Leffingwell. As the CastP technique for determining putative binding
cavities and pockets predicts the binding site for retinal in the
bovine rhodopsin models 1HXZ (chain A)5 and 1F88 (chain
A)6 with an excellent correlation to that previously
found11, CastP may provide a simple and fast
method for predicting the putative odorant binding sites in olfactory
receptors.
Background:
Previously, Pilpel & Lancet7 have inferred, that
for olfactory receptors, the odorant complementarity determining
regions reside in the transmembranal segments 3, 4, and 5.
Singer8 in an Analysis of the Molecular Basis for Octanal
Interactions in the Expressed Rat I7 Olfactory Receptor makes a
strong case that octanal binds with OR-I7 in a pocket ~10 Å
from the extracellular surface formed by transmembrane domains
3–7. In addition, Goddard, et.al.9, have
modeled the mouse receptor ORL466 (OR S25) and in docking studies
predicted the binding pocket for the compounds hexanol and heptanol.
Docking results show that TMs 3, 5, and 6 have residues directly
involved in binding and that TM4 may have an important role in
binding as it packs against TM3 and TM5 and therefore can alter their
relative position if key residues of TM4 are mutated. The presence of
a critical Lys on TM7 is similar to the related rhodopsin, where
Lys-296 (TM7) binds the retinal chromophore10 and
substitutions in this residue may switch receptor specificity toward
other functional groups.
Visuals of Receptors and Binding site
Cavities
Visual representations (derived from CastP) of the binding site
cavity in rhodopsin and the putative binding sites of a few Human
OR's of chromosone 1 are shown below.
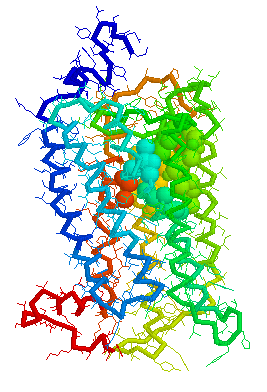
Putative Binding cavity in Human OR1.04.06 derived
using CastP
|
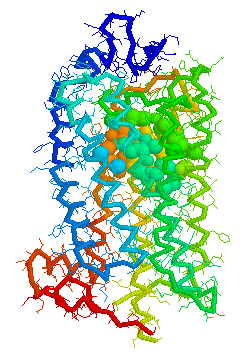
Binding cavity for retinal in
Bovine
rhodopsin 1HZX Chain A derived using CastP
|
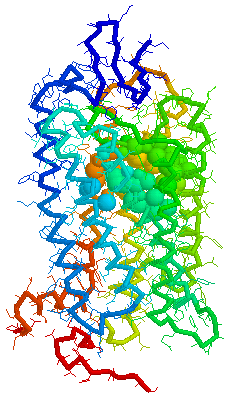
Binding cavity for retinal in
Bovine
rhodopsin 1F88 Chain A derived using CastP
|
|
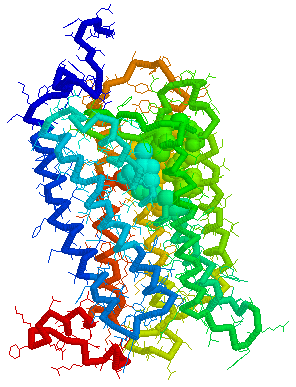
Putative Binding cavity in Human OR1.04.05 derived
using CastP
|

Putative Binding cavity in Human OR1.04.02 derived
using CastP
|

Putative Binding cavity in Human OR1.06.01 derived
using CastP
|
|
|
|
|
Sequence data used was from the work of Zozulya, Echeverri &
Nguyen of Senomyx who published a paper entitled "The human olfactory
receptor repertoire" (June, 2001) in which they reported the
identification and physical cloning of 347 putative human full-length
odorant receptor genes. Comparative sequence analysis of the
predicted gene products allowed them to identify and define a number
of consensus sequence motifs and structural features of this vast
family of receptors. They believe that these sequences represent the
essentially complete repertoire of functional human odorant
receptors.12 .They found some
differences between their data and those in the HORDE database
maintained by Doron Lancet's group at at the Weizmann Institute of
Science Crown Human Genome Center13. These included 29
"human OR" (hOR) genes that were apparently identified as pseudogenes
in the HORDE
database, but encoded as functional hOR candidates in their analysis,
as well as 10 hORs not found in HORDE. This online article includes a
downloadable file with all 347 hOR's in FASTA
format.12a
All the Human OR sequences are now available and cross referenced
to sources at the SenseLab
Olfactory Receptor DataBase (ORDB).
______________________________
PDB models of selected Human Olfactory receptors are available
from Leffingwell & Associates to interested parties upon
request.
Contact: J.C. Leffingwell at Email: leffingwell@mindspring.com
Cautionary Statement: The X-ray
crystal structures of olfactory receptors have not yet been
determined. These theoretical OR models are based on the rhodpsin
templates starting first with the OR model with best sequence fit,
with other models evolving from the first models. Models were
prepared using the Swiss-PdbViewer software as well as Swiss-Model14-16.
Certain helical areas were hand corrected to improve conformational
issues. Optimized models were checked with the WHAT
IF program at Heidelberg. As with most protein models
(theoretical or as deterimed by X-ray diffraction), these models are
not perfect as analyzed by WHAT IF. As such these models are
first approach type models and while they may be useful for
determining binding sites and in odorant docking studies, Leffingwell
& Associates does not warrant these models suitable for any
purpose.
References:
1. Edelsbrunner H, Facello M, Liang J., On
the definition and the construction of pockets in macromolecules.
Disc. Appl. Math. 88:83-102 (1998).
2. Liang J, Edelsbrunner H, Woodward C., Anatomy
of protein pockets and caviteis: measurement of binding site geometry
and implications for ligand design. Protein Science. 7:1884-1897
(1998).
3. J. Liang, H. Edelsbrunner, P. Fu, P.V. Sudhakar and S.
Subramaniam., Analytical
shape computing of macromolecules I: molecular area and volume
through alpha shape. Proteins. 33, 1-17 (1998).
4. Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S.,
Analytical
shape computation of macromolecules. II. Identification and
computation of inaccessible cavities in proteins. Proteins:
Struct. Funct. Genet. 33:18-29 (1998 ).
5. Teller, D. C., Okada, T., Behnke, C. A., Palczewski, K.,
Stenkamp, R. E.: Advances
in Determination of a High-Resolution Three-Dimensional Structure of
Rhodopsin, a Model of G-Protein-Coupled Receptors (Gpcrs)
Biochemistry 40 pp. 7761 (2001)
6. Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A.,
Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T.,
Stenkamp, R. E., Yamamoto, M., Miyano, M.: Crystal
Structure of Rhodopsin: A G Protein-Coupled Receptor Science 289
pp. 739 (2000)
7. Pilpel, Y. and D. Lancet , The
variable and conserved interfaces of modeled olfactory receptor
proteins., Protein Sci., May;8(5):969-77 (1999).
8. Singer, Michael S., Analysis
of the Molecular Basis for Octanal Interactions in the Expressed Rat
I7 Olfactory Receptor,Chem. Senses 25: 155-165, (2000)
9. Floriano WB, Vaidehi N, Goddard WA 3rd, Singer MS, Shepherd
GM., Molecular
mechanisms underlying differential odor responses of a mouse
olfactory receptor. Proc Natl Acad Sci U S A, Sep
26;97(20):10712-6 (2000).
10. Singer, M. S., Weisinger-Lewin, Y., Lancet, D. & Shepherd,
G. M., Positive
selection moments identify potential functional residues in human
olfactory receptors., Recept. Channels 4, 141-147 (1996)
11. Leffingwell, J.C., manuscript in preparation.
12. Sergey Zozulya, Fernando Echeverri & Trieu Nguyen, The
human olfactory receptor repertoire, Genome
Biology 2001 2(6): research0018.1-0018.12
12a. Zozulya, et. al., http://www.genomebiology.com/2001/2/6/research/0018/gb-2001-2-6-research0018-S1.asp
13. Fuchs T, Glusman G, Horn-Saban S, Lancet D, Pilpel Y,
The
human olfactory subgenome: from sequence to structure and
evolution, Hum Genet 2001 Jan;108(1):1-13; Glusman G, Yanai I,
Rubin I, Lancet D., The
complete human olfactory subgenome, Genome Res. 2001
May;11(5):685-702
14. Guex, N. and Peitsch, M. C., SWISS-MODEL
and the Swiss-PdbViewer: An environment for comparative protein
modelling. Electrophoresis 18:2714-2723 (1997).
15. Peitsch, M. C. ProMod
and Swiss-Model: Internet-based tools for automated comparative
protein modelling. Biochem Soc Trans 24:274-279 (1996).
16. Peitsch, M. C. Protein modeling by E-mail, Bio/Technology
13,658-660 (1995).